|
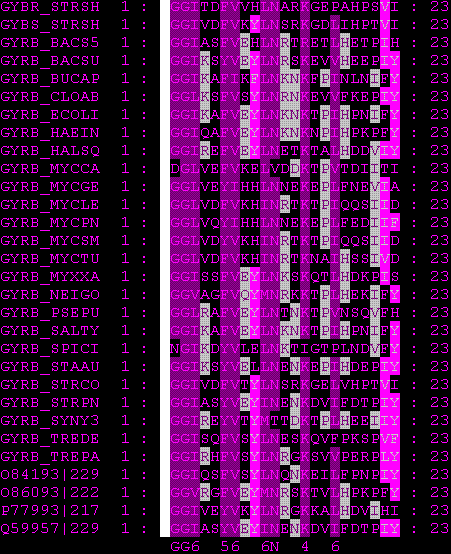
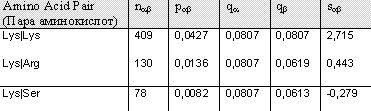
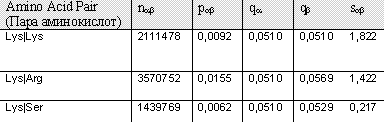
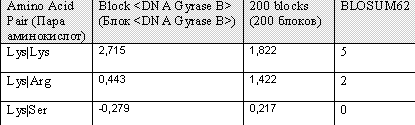
"Block DNA GyrB" shows the score of pairs Lys|Lys, Lys|Arg and Lys|Ser respectively, from a block constituted by amino acid sequences from the same protein (DNAGyraseB), but from different organisms, for example GYRB_ECOLI, GYRB_YERPE (yeast), GYRB_HEINE, GYRB_MUSC (mouse). Block 200 illustrates the score of these pairs from 200 different blocks constituted by sequences from differnet organisms. BLOSUM (BLOck SUbstitution Matrix)----. Comparison of the results shows that they badly correlate. Between the values of single BLock the differences are smaller than between the values of Blosum62. One similarity is that both matrices give the values of the three pairs in the same order of size.Analyzing the results of data from matrix shows that it correlates better with the results of data from matrix Blosum62, than what matrix Block_Pairs do; the differences between the amino acid pairs are larger in Block200_Pairs. This is probably due to that the latter is based in a wider range of alignments. Note the negative value for Lys|Ser of Block
|